Assign index peaks
assign_index_peaks.RdAssign index peaks in preparation for calculation of instability metrics
Arguments
- fragments_list
A list of "fragments" class objects representing fragment data.
- config
A trace_config object generated using
load_config().- index_override_dataframe
A data.frame to manually set index peaks. Column 1: unique sample IDs, Column 2: desired index peaks (the order of the columns is important since the information is pulled by column position rather than column name). Closest peak in each sample is selected so the number needs to just be approximate. Default:
NULL.- ...
additional parameters from any of the functions in the pipeline detailed below may be passed to this function. This overwrites values in the
config. These parameters include:groupedLogical value indicating whether samples should be grouped to share a common index peak.FALSEwill assign the sample's own modal allele as the index peak.TRUEwill use metadata to assign the index peak based on the modal peak of another sample (see below for more details). Default:FALSE.
Details
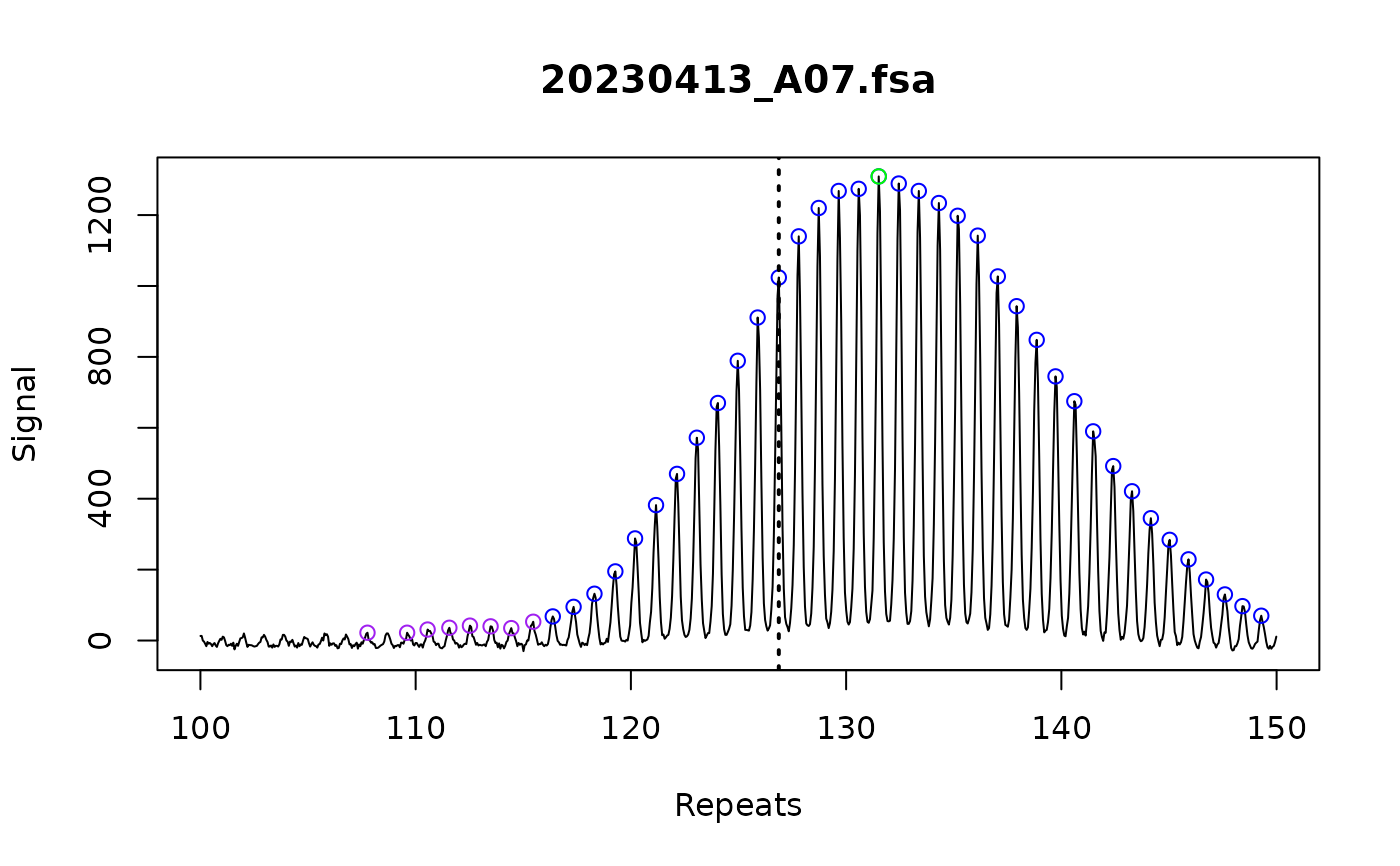
A key part of instability metrics is the index peak. This is the repeat length used as the reference peak for relative instability metrics calculations, like expansion index. This is usually the the inherited repeat length of a mouse, or the modal repeat length for the cell line at a starting time point.
If grouped is set to TRUE, this function groups the samples by their metrics_group_id and uses the samples set as metrics_baseline_control to set the index peak. Use add_metadata() to set these variables. This is useful for cases like inferring repeat size of inherited alleles from mouse tail data. If the samples that are going to be used to assign index peak are from different fragment analysis runs, use correction = "batch" in call_repeats() to make sure the systematic differences between runs are corrected and the correct index peak is assigned. If there are multiple samples used as baseline control, the median value will be used to assign index peak to corresponding samples.
For mice, if just a few samples have the inherited repeat signal shorter than the expanded population, you could not worry about this and instead use the index_override_dataframe. This can be used to manually override these assigned index repeat values (irrespective of whether grouped is TRUE or FALSE).
As a final option, the index peak could be manually assigned directly to a fragments class using the internal setter function fragments$set_index_peak().
Examples
fsa_list <- lapply(cell_line_fsa_list, function(x) x$clone())
config <- load_config()
trace:::add_metadata(
fsa_list,
metadata_data.frame = trace::metadata
)
trace:::find_ladders(fsa_list, config, show_progress_bar = FALSE)
trace:::find_fragments(fsa_list, config,
min_bp_size = 300,
show_progress_bar = FALSE
)
trace:::find_alleles(
fsa_list,
config
)
trace:::call_repeats(
fsa_list,
config
)
trace:::assign_index_peaks(
fsa_list,
config,
grouped = TRUE
)
plot_traces(fsa_list[1], xlim = c(100,150))